2025
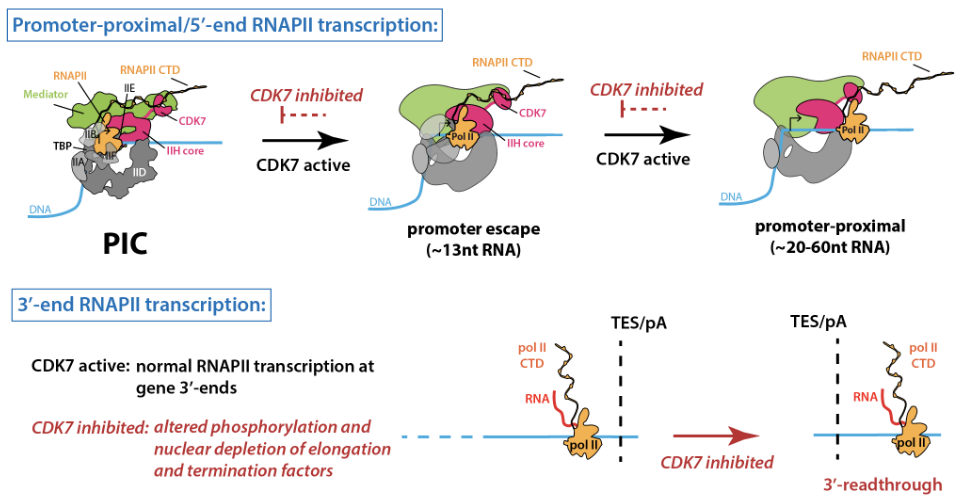
TFIIH kinase CDK7 drives cell proliferation through a common core transcription factor network
T. Jones, J. Feng, O. Luyties, K. Cozzolino, L. Sanford, J.K. Rimel, C.C. Ebmeier, G.S. Shelby, L.P. Watts, J. Rodino, N. Rajagopal, S. Hu, F. Brennan, Z.L. Maas, S. Alnemy, W.F. Richter, A.F. Koh, N.B. Cronin, A. Madduri, J. Das, E. Cooper, K.B. Hamman, J.P. Carulli, M.A. Allen, S. Spencer, A. Kotecha, J.J. Marineau, B.J. Greber, R.D. Dowell, D.J. Taatjes. Scientific Advances 11(9):eadr9660 (2025)
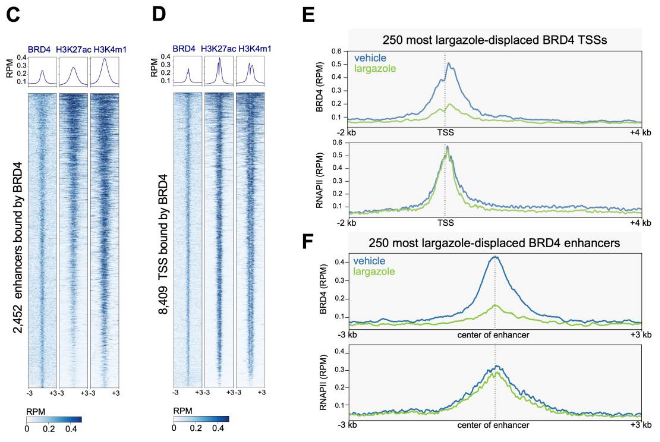
Histone Deacetylase Inhibitor Largazole Deactivates A Subset of Superenchancers and Causes Mitotic Chromosome Mis-alignment by Suppressing SP1 and BRD4
G.J. Sanchez, Z. Liu, S. Hunter, Q. Xu, J.T.V. Westfall, G.E. Wheeler, C. Toomey, D. Taatjes, M. Allen, R.D. Dowell, X. Liu. bioRxiv (2025)
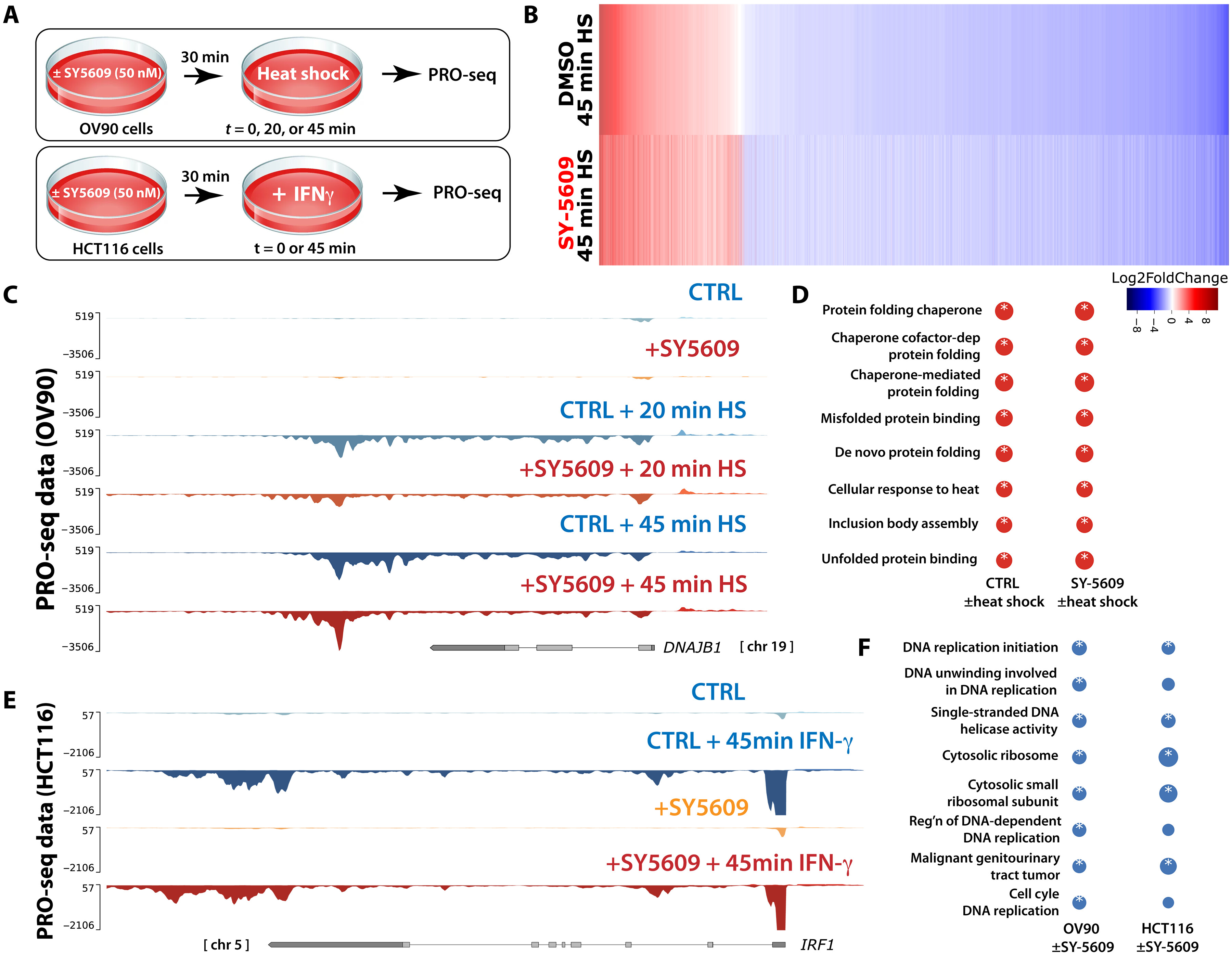
Multi-omics and biochemical reconstitution reveal CDK7-dependent mechanisms controlling RNA polymerase II function at gene 5'- and 3'-ends
O. Luyties, L. Sanford, J. Rodino, M. Nagel, T. Jones, J.K. Rimel, C.C. Ebmeier, G.S. Shelby, K. Cozzolino, F. Brennan, A. Hartzog, M.B. Saucedo, L.P. Watts, S. Spencer, J.F. Kugel, R.D. Dowell, D.J. Taatjes. bioRxiv (2025)
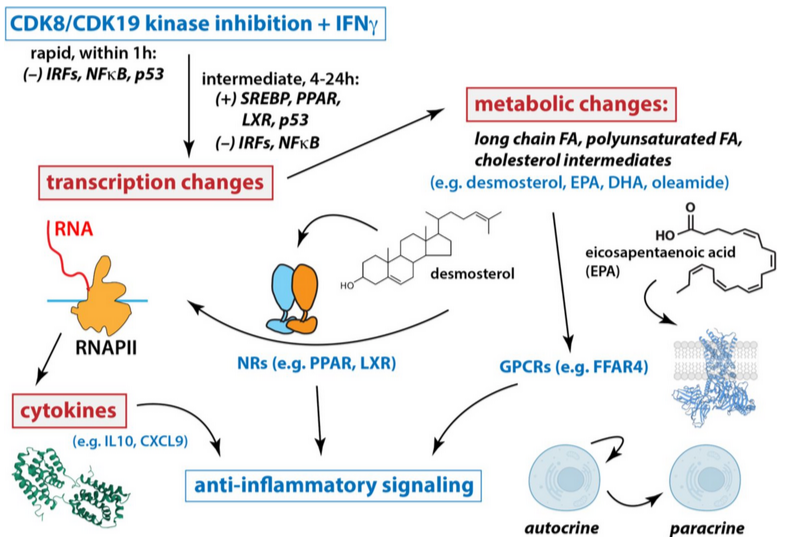
Mediator kinase inhibition suppresses hyperactive interferon signaling in Down syndrome
K. Cozzolino, L. Sanford, S. Hunter, K. Molison, B. Erickson, T. Jones, M.C.S. Courvan, D. Ajit, M.D. Galbraith, J.M. Espinosa, D.L. Bentley, M.A. Allen, R.D. Dowell, D.J. Taatjes. eLife 13:RP100197 (2025)
2024
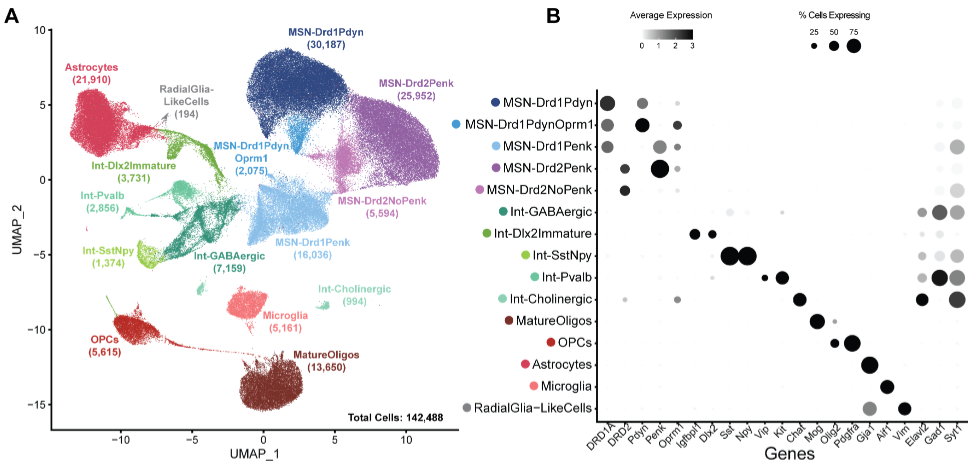
Single nucleus RNA-sequencing reveals transcriptional synchrony across different relationships
L.E. Brusman, J.M. Sadino, A.C. Fultz, M.A. Kelberman, R.D. Dowell, M.A. Allen, Z.R. Donaldson. bioRxiv (2024)
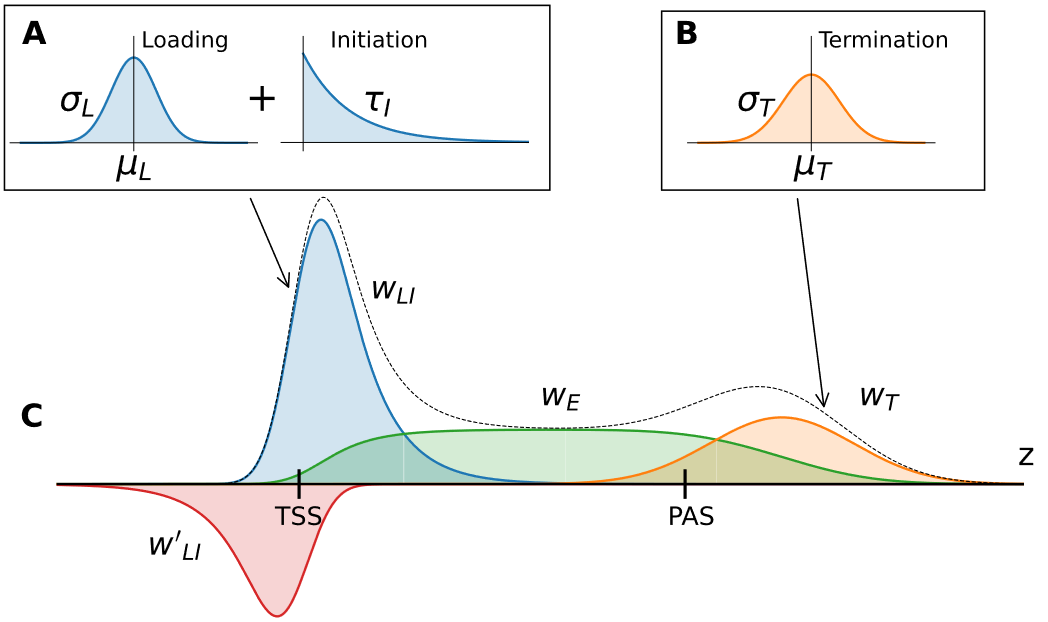
LIET Model: Capturing the kinetics of RNA polymerase from loading to termination
J. Stanley, G.E.F. Barone, H.A. Townsend, R.F. Sigauke, M.A. Allen, R.D. Dowell. bioRxiv (2024)
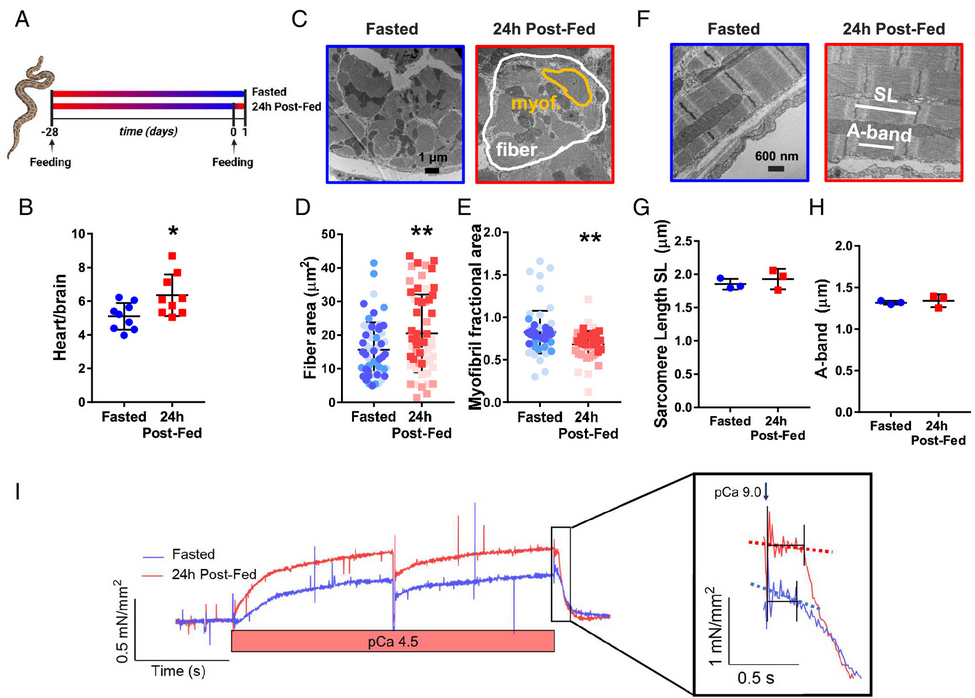
Postprandial cardiac hypertrophy is sustained by mechanics, epigenetic, and metabolic reprogramming in pythons
C. Crocini, K.C. Woulfe, C.D. Ozeroff, S. Perni, J. Cardiello, C.J. Walker, C.E. Wilson, K. Anseth, M.A. Allen, L.A. Leinwand. PNAS 121 (36) e2322726121 (2024)
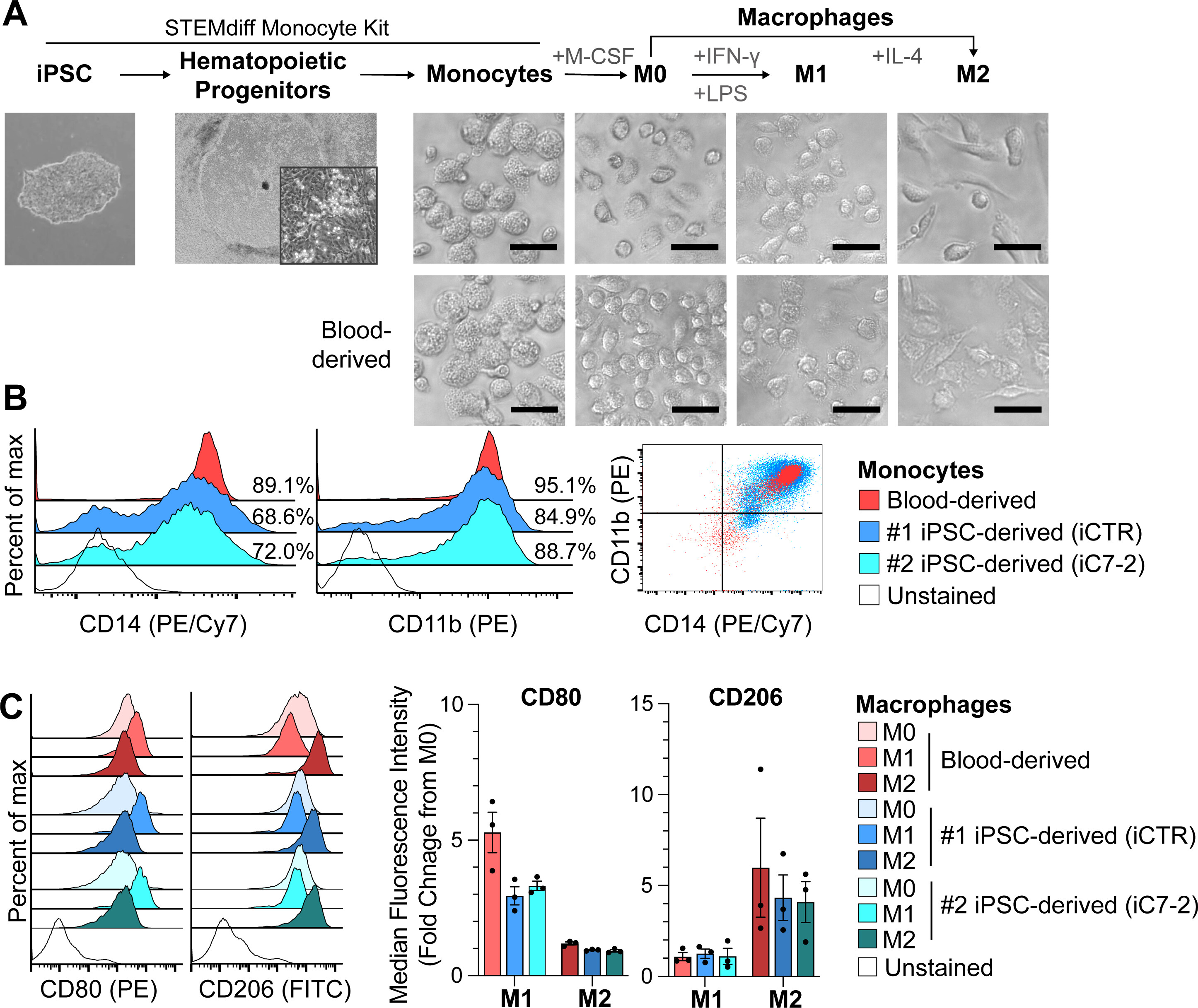
Macrophages derived from human induced pluripotent stem cells (iPSCs) serve as a high-fidelity cellular model for investigating HIV-1, dengue, and influenza viruses
Q. Yang, A. Barbachano-Guerrero, L.M. Fairchild, T.J. Rowland, R.D. Dowell, M.A. Allen, C.J. Warren, S.L. Sawyer. Journal of Virology 98:e01563-23 (2024)
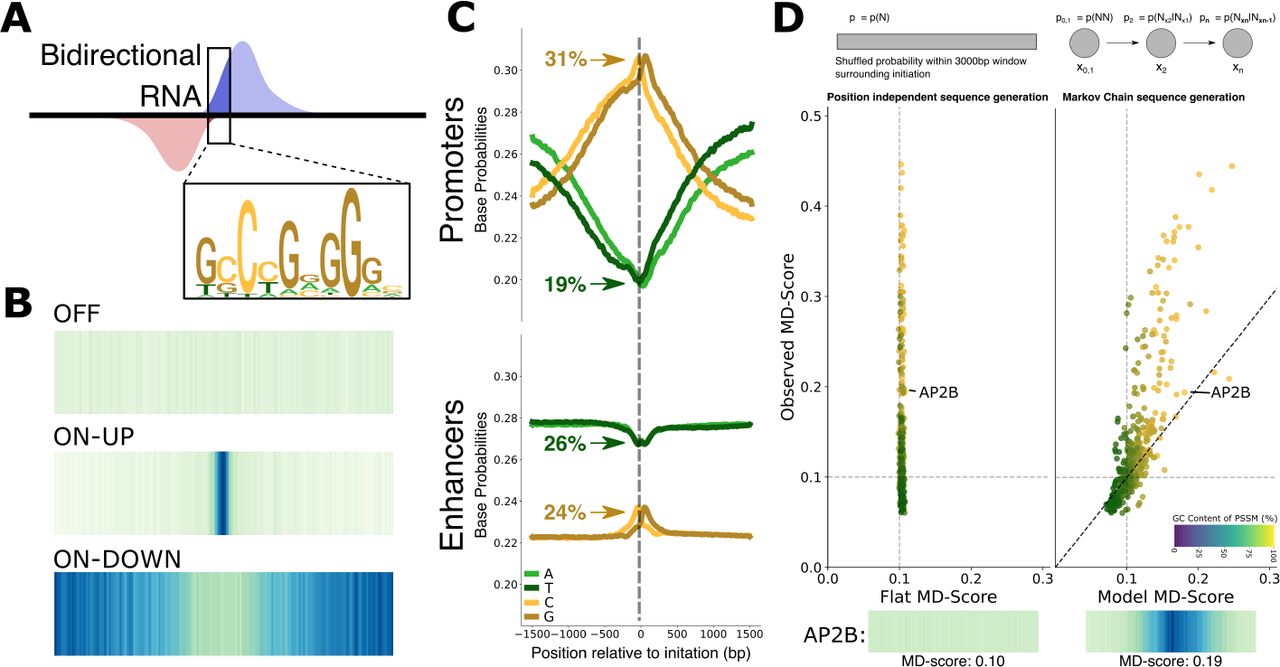
A transcription factor (TF) inference method that broadly measures TF activity and identifies mechanistically distinct TF networks
T. Jones, R.F. Sigauke, L. Sanford, D.J. Taatjes, M.A. Allen, R.D. Dowell. bioRxiv (2024)
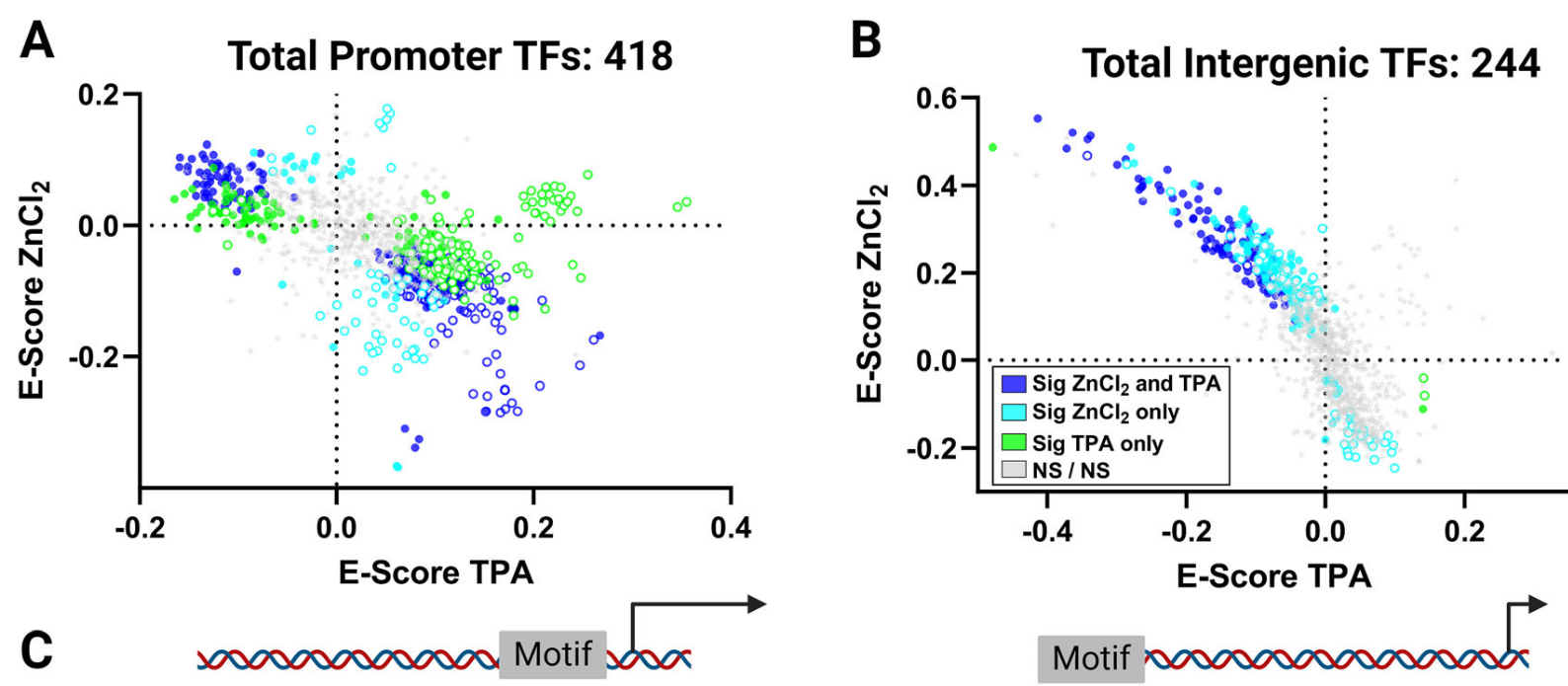
Cellular zinc status alters chromatin accessibility and binding of p53 to DNA
D. Ocampo, L.J. Damon, L. Sanford, S.E. Holtzen, T. Jones, M.A. Allen, R.D. Dowell, A.E. Palmer. Life Science Alliance 7, 9 (2024)

The temporal dynamics of lncRNA Firre-mediated epigenetic and transcriptional regulation
C. Much, E.L. Lasda, I.T. Pereira, T.K. Vallery, D. Ramirez, J.P. Lewandowski, R.D. Dowell, M.J. Smallegan, J.L. Rinn. Nature Communications 15, Article number: 6821 (2024)

Internal and external normalization of nascent RNA sequencing run-on experiments
Z. Maas, R. Dowell. BMC Bioinformatics 25, Article number: 19 (2024)

Characterizing primary transcriptional responses to short term heat shock in Down syndrome
J.F. Cardiello, J. Westfall, R. Dowell, M.A. Allen. PLoS ONE 19(8):e0307375 (2024)
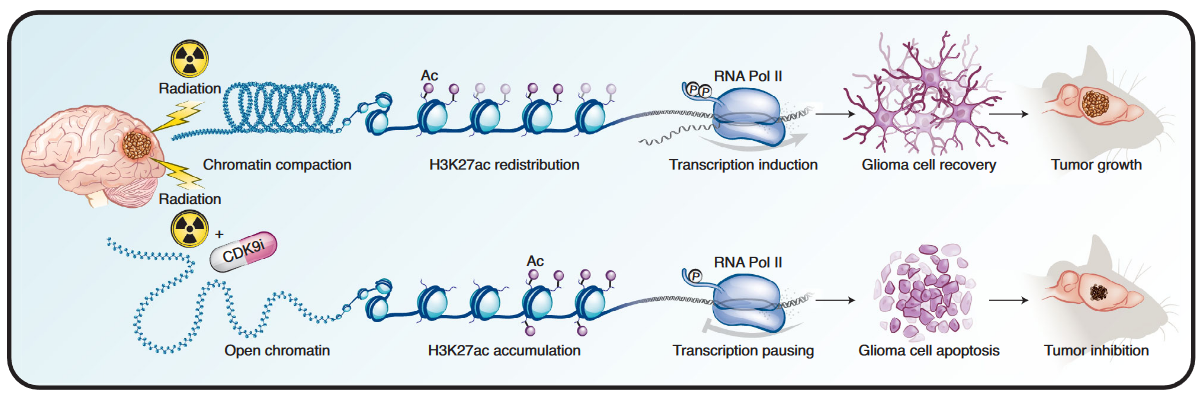
Rapid PTEFb-dependent transcriptional reorganization underpins the glioma adaptive response to radiotherapy
F.M. Walker, L.M. Sobral, E. Danis, B. Sanford, S. Donthula, I. Balakrishnan, D. Wang, A. Pierce, S.D. Karam, S. Kargar, N.J. Serkova, N.K. Foreman, S. Venkataraman, R. Dowell, R. Vibhakar, N.A. Dahl. Nature Communications 15, Article number: 4616 (2024)
2023

Deconvolution of Nascent Sequencing Data Using Transcriptional Regulatory Elements
Z. Maas, R. Sigauke, R. Dowell. Biocomputing (2023)

Noncoding SNPs decrease expression of FABP5 during COPD exacerbations
M. El Kharbili, S.K. Sasse, L. Sanford, S. Jacobson, K. Aviszus, A. Gupta, C.J. Guo, S.M. Majka, R.D. Dowell, A.N. Gerber, R.P. Bowler, F. Gally. Journal of Clinical Investigation (2023)

Atlas of nascent RNA transcripts reveals enhancer to gene linkages
R.F. Sigauke, L. Sanford, Z.L. Maas, T. Jones, J.T. Stanley, H.A. Townsend, M.A. Allen, R.D. Dowell. bioRxiv (2023)

RUN(X) out of blood: emerging RUNX1 functions beyond hematopoiesis and links to Down syndrome
E. Rozen, C.D. Ozeroff, M.A. Allen. Human Genomics 17, Article number: 83 (2023)

An intronic LINE-1 regulates IFNAR1 expression in human immune cells
C.A. Buttler, D. Ramirez, R.D. Dowell, E.B. Chuong. Mobile DNA 14, Article number: 20 (2023)

Determination of steady-state transcriptome modifications associated with repeated homotypic stress in the rat rostral posterior hypothalamic region
S. Campeau, C. McNulty, J.T. Stanley, A.N. Gerber, S.K. Sasse, R.D. Dowell. Frontiers in Neuroscience Volume 17 (2023)

Transcription dosage compensation does not occur in Down syndrome
S. Hunter, J. Hendrix, J. Freeman, R.D. Dowell, M.A. Allen. BMC Biology 21, Article number: 228 (2023)

Single cell analysis of transcriptome and open chromatin reveals the dynamics of hair follicle stem cell aging
C. Zhang, D. Wang, R. Dowell, R. Yi. Frontiers in Aging Volume 4 (2023)

Human mRNA in saliva can correctly identify individuals harboring acute infection
Q. Yang, N.R. Meyerson, C.L. Paige, J.H. Morrison, S.K. Clark, W.T. Fattor, C.J. Decker, H.R. Steiner, E. Lian, D.B. Larremore, R. Perera, E.M. Poeschla, R. Parker, R.D. Dowell, S.L. Sawyer. mBio e0171223 (2023)

Nascent transcription upon interferon-α2 stimulation on human and rhesus macaque lymphoblastoid cell lines
D. Ramirez, E.B. Chuong, R.D. Dowell. BMC Research Notes 16, Article number: 292 (2023)

Aging disrupts gene expression timing during muscle regeneration
J.V. Kurland, A.A. Cutler, J.T. Stanley, N.D. Betta, A. Van Deusen, B. Pawlikowski, M. Hall, T. Antwine, A. Russell, M.A. Allen, R. Dowell, B. Olwin. Stem Cell Reports Volume 18, Issue 6 (2023)
2022

Glucocorticoid-regulated bidirectional enhancer RNA transcription pinpoints functional genetic variants linked to asthma
S.K. Sasse, A. Dahlin, L. Sanford, M.A. Gruca, A. Gupta, A. Chen Wu, C. Ibarren, R.D. Dowell, S.T. Weiss, A.N. Gerber. medRxiv (2022)

Extracellular matrix stiffness controls cardiac valve myofibroblast activation through epigenetic remodeling
C.J. Walker, D. Batan, C.T. Bishop, D. Ramirez, B.A. Aguado, M.E. Schroeder, C. Crocini, J. Schwisow, K. Moulton, L. Macdougall, R.M. Weiss, M.A. Allen, R. Dowell, L.A. Leinwand, K.S. Anseth. BioEngineering and Translational Medicine 7(3):e10394 (2022)

Integrated genomics approaches identify mediators of transcriptional and epigenetic responses to Afghan desert particulate matter in human small airway epithelial cells
A. Gupta, S.K. Sasse, R. Berman, M.A. Gruca, R.D. Dowell, H.W. Chu, G.P. Downey, A.N. Gerber. Physiological Genomics 54(10):389-401 (2022)

Protocol Variations in Run-On Transcription Dataset Preparation Produce Detectable Signatures in Sequencing Libraries
S. Hunter, R. Sigauke, J. Stanley, M. Allen, R.D. Dowell. BMC Genomics 23, 187 (2022)

Suppression of p53 response by targeting p53-Mediator binding with a stapled peptide
B.L. Allen*, K. Quach*, T. Jones*, C.B. Levandowski, C.C. Ebmeier, J.D. Rubin, T. Read, R.D. Dowell, A. Schepartz, D.J. Taatjes. Cell Rep. 39(1):110630 (2022)
2021

Deconvolution of multiplexed transcriptional responses to wood smoke particles defines rapid aryl hydrocarbon receptor signaling dynamics
A. Gupta, S.K. Sasse, M.A. Gruca, L. Sanford, R.D. Dowell, A.N. Gerber. J. of Biol. Chemistry 297(4):101147 (2021)

Escape of hair follicle stem cells causes stem cell exhaustion during aging
C. Zhang, D. Wang, J. Wang, L. Wang, W. Qiu, T. Kume, R. Dowell, and R. Yi. Nature Aging 1,889-903 (2021)

Characterization of global gene expression, regulation of metal ions, and infection outcomes in immune competent 129S6 mouse macrophages
L.N. Janiszewski, M. Minson, M.A. Allen, R.D. Dowell, A.E. Palmer. Infect. Immun. (2021)

Chromatin remodeling due to degradation of citrate carrier impairs osteogenesis of aged mesenchymal stem cells
A. Pouikli, S. Parekh, M. Maleszewska, M. Baghdadi, I. Tripodi, C. Nikopoulou, K. Folz-Donahue, Y. Hinze, A. Mesaros, P. Giavalisco, R. Dowell, L. Partridge, and P. Tessarz. Nature Aging 1,810-825 (2021)

The Δ40p53 isoform inhibits p53-dependent eRNA transcription and enables regulation by signal-specific transcription factors during p53 activation
C.B. Levandowski, T. Jones, M. Gruca, S. Ramamoorthy, R.D. Dowell#, D.J. Taatjes#. PLoS Biology 19(8): e3001364 (2021)

Application of a bioinformatic pipeline to RNA-seq data identifies novel viruslike sequence in human blood
M. Melnick, P. Gonzales, T.J. LaRocca, Y. Song, J. Wuu, M. Benatar, B. Oskarsson, L. Petrucelli, R.D. Dowell, C.D. Link, M. Prudencio. G3 11:9 (2021)

Just 2% of SARS-CoV-2-positive individuals carry 90% of the virus circulating in communities
Q. Yang, T.K. Saldi, E. Lasda, C.J. Decker, C.L. Paige, D. Muhlrad, P.K. Gonzales, M.R. Fink, K.L. Tat, C.R. Hager, J.C. Davis, C.D. Ozeroff, N.R. Meyerson, S.K. Clark, W.T. Fattor, A.R. Gilchrist, A. Barbachano-Guerrero, E.R. Worden-Sapper, S.S. Wu, G.R. Brisson, M.B. McQueen, R.D. Dowell, L. Leinwand, R. Parker, S.L. Sawyer. PNAS 118(21):e2104547118 (2021)

Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts
C.J. Walker, C. Crocini, D. Ramirez, A.R. Killaars, J.C. Grim, B.A. Aguado, S.K. Clark, M. Allen, R. Dowell, L.A. Leinwand*, K.S. Anseth*. Nature Biomedical Engineering 5, 1517-1518 (2021)

Competition between PRC2.1 and 2.2 subcomplexes regulates PRC2 chromatin occupancy in human stem cells
D.T. Youmans, A.R. Gooding, R.D. Dowell, T.R. Cech. Molecular Cell 81(3):p488-501.E9 (2021)

The MUC5B-associated variant rs35705950 resides within an enhancer subject to lineage- and disease-dependent epigenetic remodeling
F. Gally, S.K. Sasse, J.S. Kurche, M.A. Gruca, J.H. Cardwell, T. Okamoto, H.W. Chu, X. Hou, O.B. Poirion, J. Buchanan, S. Preissl, B. Ren, S.P. Colgan, R.D. Dowell, I.V. Yang, D.A. Schwartz, A.N. Gerber. JCI Insight 6(2) (2021)

Transcription factor enrichment analysis (TFEA) quantifies the activity of multiple transcription factors from a single experiment.
J.D. Rubin, J.T. Stanley, R.F. Sigauke, C.B. Levandowski, Z.L. Maas, J. Westfall, D.J. Taatjes & R.D. Dowell. Communications Biology 4:661 (2021)
Earlier Publications
Systems genetics analysis of the LXS recombinant inbred mouse strains: Genetic and molecular insights into acute ethanol tolerance
R.A. Radcliffe , R. Dowell, A.T. Odell, P.A. Richmond, B. Bennett, C. Larson, K. Kechris, L.M. Saba, P. Rudra, S. Wen.
PLoS ONE 15(10): e0240253 (2020)
Selective inhibition of CDK7 reveals high-confidence targets and new models for TFIIH function in transcription
J.K. Rimel, Z.C. Poss, B. Erickson, Z.L. Maas, C.C. Ebmeier, J.L. Johnson, T-M. Decker, T.M. Yaron, M.J. Bradley, K.B. Hamman, S. Hu, G. Malojcic, J.J. Marineau, P.W. White, M. Brault, L. Tao, P. DeRoy, C. Clavette, S. Nayak, L.J. Damon, I.H. Kaltheuner, H. Bunch, L.C. Cantley, M. Geyer, J. Iwasa, R.D. Dowell, D.L. Bentley, W.M. Old and D.J. Taatjes.
Genes and Development 34: 1452-1473 (2020)
Applying knowledge-driven mechanistic inference to toxicogenomics
I.J. Tripodi, T. Callahan, J. Westfall, N. Meitzer, R. Dowell, L. Hunter.
Toxicology in Vitro 66:104877 (2020)
Combining signal and sequence to detect RNA polymerase initiation in ATAC-seq data
I.J. Tripodi, M. Chowdhury, M. Gruca, R.D. Dowell.
PLoS ONE 15(4):e0232332 (2020)
TFIID enables RNA Polymerase II promoter-proximal pausing
C.B. Fant, C.B. Levandowski, K. Gupta, Z.L. Maas, J. Moir, J.D. Rubin, A. Sawyer, M.N. Esbin, J.K. Rimel, O. Luyties, M.T. Marr, I. Berger, R.D. Dowell, D.J. Taatjes.
Molecular Cell 78(4):785-793 (2020)
Remodeling of Zn2+ homeostasis upon differentiation of mammary epithelial cells
Y. Han, L. Sanford, D.M. Simpson, R.D. Dowell, A.E. Palmer.
Metallomics 12:346-362 (2020)
Lessons from eRNAs: understanding transcriptional regulation through the lens of nascent RNAs.
J.F. Cardiello, G.J. Sanchez, M.A. Allen, R.D. Dowell.
Transcription 11(1):3-18 (2020)
Two-stage ML Classifier for Identifying Host Protein Targets of the Dengue Protease
J.T. Stanley, A.R. Gilchrist, A.C. Stabell, M.A. Allen, S.L. Sawyer, and R.D. Dowell.
Pac Symp Biocomputing 2020 Dec 2019, 487-498 (2019)
Annotation Agnostic Approaches to Nascent Transcription Analysis: Fast Read Stitcher and Transcription Fit
M.A. Gruca, M.A. Gohde, R.D. Dowell.
Methods in Molecular Biology (2019)
Nascent transcript analysis of glucocorticoid crosstalk with TNF defines primary and cooperative inflammatory repression
S.K. Sasse, M. Gruca, M.A. Allen, V. Kadiyala, T. Song, F. Gally, A. Gupta, M.A. Pufall, R.D. Dowell, A.N. Gerber.
Genome Research 29:1753-1765 (2019)
Transcriptional Responses to INFgamma require mediator kinase-dependent pause release and mechanistically distinct CDK8 and CDK19 functions
I. Steinparzer, V. Sedlyarov, J.D. Rubin, K. Eislmayr, M.D. Galbraith, C.B. Levandowski, T. Vcelkova, L. Sneezum, F. Wascher, F. Amman, R. Kleinova, J.M. Espinosa, G. Superti-Furga, R.D. Dowell, D.J. Taatjes*, P. Kovarik*.
Molecular cell 76(3):485-499 (2019)
Identification and characterization of a novel anti-inflammatory lipid isolated from Mycobacterium vaccae, a soil-derived bacterium with immunoregulatory and stress resilience properties
D.G. Smith, R. Martinelli, G.S. Besra, P.A. Illarionov, I. Szatmari, P. Brazda, M.A. Allen, W. Xu, X. Wang, L. Nagy, R.D. Dowell, G.A. W. Rook, L.R. Brunet, C.A. Lowry.
Psychopharmacology May;236(5):1653-1670 (2019)
Heat shock in C. elegans induces downstream of gene transcription and accumulation of double-stranded RNA
M. Melnick, P. Gonzales, J. Cabral, M.A. Allen, R.D. Dowell, C.D. Link.
PLoS ONE Apr 8;14(4):e0206715 (2019)
Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation.
S. Cuartero, F.D. Weiss, G. Dharmalingam, Y. Guo, E. Ing-Simmons, S. Masella, I. Robles-Rebollo, X. Xiao, Y.F. Wang, I. Barozzi, D. Djeghloul, M.T. Amano, H. Niskanen, E. Petretto, R.D. Dowell, K. Tachibana, M.U. Kaikkonen, K.A. Nasmyth, B. Lenhard, G. Natoli, A.G. Fisher, and M. Merkenschlager.
Nature Immunology Sept 19(9):932-941 (2018)
PI(3,5)P2 Controls Vacuole Potassium Transport to Support Cellular Osmoregulation
Z.N. Wilson, A.L. Scott, R.D. Dowell, and G. Odorizzi.
Molecular Biology of the Cell 29(14):1718-1731 (2018)
Detecting Differential Transcription Factor Activity from ATAC-Seq Data
I.J. Tripodi, M.A. Allen, and R.D. Dowell.
Molecules 23(5), 1136 (2018)
miR-MaGiC improves quantification accuracy for small RNA-seq
P.H. Russell, B. Vestal, W. Shi, P.D. Rudra, R. Dowell, R. Radcliffe, L. Saba and K. Kechris.
BMC Research Notes 11:296 (2018)
Enhancer RNA profiling predicts transcription factor activity
J.G Azofeifa, M.A. Allen, J.R. Hendrix, T. Read, J.D. Rubin, and R.D. Dowell.
Genome Research 28: 334-344 (2018)
Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers
G.J. Sanchez, P.A. Richmond, E.N. Bunker, S.S. Karman, J. Azofeifa, A.T. Garnett, Q. Xu, G.E. Wheeler, C.M. Toomey, Q. Zhang, R.D. Dowell and X. Liu.
Nucleic Acids Research 46(4):1756-1776 (2018)
A communal catalogue reveals Earth’s multiscale microbial diversity
L.R. Thompson, J.G. Sanders, D. McDonald, A. Amir, J. Ladau, K.J. Locey, ..(et. al.) .. and The Earth Microbiome Project Consortium.
Nature 551(7681):457-463 (2017)
A kinase-independent role for CDK19 in p53 response
K.A. Audetat, M.D. Galbraith, A.T. Odell, T. Lee, A. Pandey, J.M. Espinosa, R.D. Dowell and D.J. Taatjes.
Molecular and Cellular Biology 37:13 (2017)
Discriminating between HuR and TTP binding sites using the k-spectrum kernel method.
S. Bhandare, D.S. Goldberg, and R.D. Dowell.
PLoS ONE 12(3):e0174052 (2017)
Transcriptome and Functional Profile of Cardiac Myocytes Is Influenced by Biological Sex
C.L. Trexler, A.T. Odell, M.Y. Jeong, R.D. Dowell, L.A. Leinwand.
Circulation: Cardiovascular Genetics 10:e001770 (2017)
Human TFIIH kinase CDK7 regulates transcription-associated epigenetic modifications
C.C. Ebmeier, B. Erickson, B.L. Allen, M.A. Allen, H. Kim, N. Fong, J.R. Jacobsen, K. Liang, A. Shilatifard, R.D. Dowell, W.M. Old, D.L. Bentley, and D.J. Taatjes.
Cell Reports 20(5):1173-1186 (2017)
The Influence of Polyploidy on the Evolution of Yeast Grown in a Sub-Optimal Carbon Source
A.L. Scott, P.A. Richmond, R.D. Dowell, A.M. Selmecki.
Molecular Biology and Evolution 34(10):2690-2703 (2017)
A generative model for the behavior of RNA polymerase
J.G. Azofeifa and R.D. Dowell.
Bioinformatics 33(2):227-234 (2017)
Model based heritability scores for high-throughput sequencing data
P. Rudra, W. J. Shi, B. Vestal, P.H. Russell, A. Odell, R.D. Dowell, R.A. Radcliffe, L.M. Saba and K. Kechris.
BMC Bioinformatics 18:143 (2017)
RNA Pol II transcription model and interpretation of GRO-seq data
M.E. Lladser, J.G. Azofeifa, M.A. Allen, R.D. Dowell.
Journal of Mathematical Biology 74:77 (2017)
An annotation agnostic algorithm for detecting nascent RNA transcripts in GRO-seq
J. Azofeifa, M.A. Allen, M.Lladser, and R.D. Dowell.
IEEE/ACM Transactions on Computational Biology and Bioinformatics 14(5):1070-1081 (2017)
Genome Characterization of the Selected Long and Short Sleep Mouse Lines
R.D. Dowell, A. Odell, P. Richmond, D. Malmer, E. Halper-Stromberg, B. Bennett, C. Larson, S. Leach, R.A. Radcliffe.
Mammalian Genome 27:574 (2016)
Survey of cryptic unstable transcripts in yeast
J. Vera and R.D. Dowell.
BMC Genomics 17:305 (2016)
Evidence for Association Between Low Frequency Variants in CHRNA6/CHRNB3 and Antisocial Drug Dependence
H.M. Kamens, R.P. Corley, P.A. Richmond, T.M. Darlington, R.Dowell, C.J. Hopfer, M.C. Stallings, J.K. Hewitt, S.A. Brown, M.A. Ehringer.
Behavior Genetics Sep;46(5):693-704 (2016)
Identification of Mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics.
Z.C. Poss, C.C. Ebmeier, A. Odell, A. Tangpeerachaikul, T. Lee, H. E. Pelish, M.D. Shair, R.D. Dowell, W. Old, D.J. Taatjes.
Cell Reports Apr 12;15(2):436-50 (2016)
Foxc1 reinforces quiescence in self-renewing hair follicle stem cells.
L. Wang, J.A. Siegenthaler, R.D. Dowell, R. Yi.
Science 351(6273): 613-617 (2016)
A trans-acting variant within the transcription factor RIM101 interacts with genetic background to determine its regulatory capacity
T. Read, P.A. Richmond, and R.D. Dowell.
PLoS Genetics 12(1): e1005746 (2016)
A modeling framework for generation of positional and temporal simulations of transcriptional regulation
D.A. Knox and R.D. Dowell.
IEEE/ACM Transactions on Computational Biology and Bioinformatics May-June 13(3):459-71; ISSN: 1545-5963 (2016)
Prolonged Cre expression driven by the α-myosin heavy chain promoter can be cardiotoxic
E.K. Pugach, P.A. Richmond, J.G. Azofeifa, R.D. Dowell, L.A. Leinwand.
Journal of Molecular and Cellular Cardiology 86:54-61 (2015)
Polyploidy can drive rapid adaptation in yeast
A. Selmecki, Y.E. Maruvka, P.A. Richmond, M. Gullet, N. Shoresh, A. Sorenson, S. De, R. Kishony, F. Michor, R.D. Dowell, D. Pellman.
Nature 519, 349–352 (2015)
QTL mapping of acute functional tolerance in the LXS recombinant inbred strains
B. Bennett, C. Larson, P.A. Richmond, A. Odell, L. Saba, B. Tabakoff, R.D. Dowell, R. Radcliffe.
Alcoholism: Clinical and Experimental Research 39(4):611-620 (2015)
An Engineered Calcium-Precipitable Restriction Enzyme
J. Hendrix, T. Read, J-F. Lalonde, P.K Jensen, W. Heymann, E.Z. Lovelace, S.A. Zimmermann, M. Brasino, J. Rokicki, R.D. Dowell.
ACS Synthetic Biology 3(12):969-971 (2014)
Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms
M.A. Allen, Z. Andrysik, V.L. Dengler, H.S. Mellert, A. Guarnieri, J.A. Freeman, K.D. Sullivan, M.D. Galbraith, X. Luo, W.L. Kraus, R.D. Dowell, and J.M. Espinosa.
eLife 2014;3:e02200 (2014)
CodaChrome: a tool for the visualization of proteome conservation across all fully sequenced bacterial genomes
J. Rokicki, D. Knox, R.D. Dowell, and S.D. Copley.
BMC Genomics 15:65 (2014)
FStitch: A fast and simple algorithm for detecting nascent RNA transcripts
J. Azofeifa, M.A. Allen, M.Lladser, and R.D. Dowell.
5th ACM Conference on Bioinformatics, Computational Biology and Health Informatics Sept. (2014)
Retrospective reflections of a whistleblower - Opinions on misconduct responses
M.A. Allen and R.D. Dowell.
Accountability in Research 20:5-6, 339-348 (2013)
HIF1A employs CDK8-Mediator to stimulate RNAPII elongation in response to hypoxia
M.D. Galbraith, M.A. Allen, C.L. Bensard, X. Wang, M.K. Schwinn, B. Qin, H.W. Long, D.L. Daniels, W.C. Hahn, R.D. Dowell and J.M. Espinosa.
Cell 153(6) pp. 1327 - 1339 (2013)
Genome-wide maps of polyadenylation reveal dynamic mRNA 3′-end formation in mammalian cell lineages
L. Wang, R.D. Dowell and R. Yi.
RNA 19(3):413-425 (2013)
sizzled function and secreted factor network dynamics
J. Shi, H. Zhang, R.D. Dowell and M.W. Klymkowsky.
Biology Open 1, 286-294 (2012)
Discovering Regulatory Overlapping RNA Transcripts
T. Danford, R. Dowell, S. Agarwala, P. Grisafi, G. Fink, D. Gifford.
Journal of Computational Biology 18(3): 295-303 (2011)
The similarity of gene expression between human and mouse tissues
R.D. Dowell.
Research Highlight - Genome Biology 12:101 (2011)
Transcription factor binding variation in the evolution of gene regulation
R.D. Dowell.
Trends in Genetics 26(11):468-475 (2010)
Regulatory Overlapping RNA Transcripts
T. Danford, R. Dowell, S. Agarwala, P. Grisafi, G. Fink, D. Gifford.
14th Annual International Conference, RECOMB LNBI 6044:110-122 (2010)
Feed-forward Regulation of a Cell Fate Determinant by an RNA-binding Protein Generates Asymmetry in Yeast
J.J. Wolf, R.D. Dowell, S. Mahony, M. Rabani, D.K. Gifford, G.R. Fink.
Genetics 185:513-522 (2010)
Genotype to Phenotype: A Complex Problem
R.D. Dowell*, O. Ryan*, A. Jansen, D. Cheung, S. Agarwala, T. Danford, D.A. Bernstein, P.A. Rolfe, L.E. Heisler,B. Chin, C. Nislow, G. Giaever, P.C. Phillips, G.R. Fink, D.K. Gifford, C. Boone.
Science 328(5977):469 (2010)
Toggle involving cis-interfering RNAs controls variegated gene expression in yeast
S.L. Bumgarner, R.D. Dowell, P. Grisafi, D.K. Gifford, G.R. Fink.
PNAS 106(43):18321-18326 (2009)
Automated discovery of functional generality of human gene expression programs
G.K. Gerber, R.D. Dowell, T.S. Jaakkola, and D.K. Gifford.
PLoS Computational Biology 3(8):e148 (2007)
Tissue-specific transcriptional regulation has diverged significantly between human and mouse
D.T. Odom, R.D. Dowell, E.S. Jacobsen, W. Gordon, T.W. Danford, K.D. MacIsaac, P.A. Rolfe, C.M. Conboy, D.K. Gifford, and E. Fraenkel.
Nature Genetics 39(6):730-2. (2007)
Efficient pairwise RNA structure prediction and alignment using sequence alignment constraints
R.D. Dowell and S.R. Eddy.
BMC Bioinformatics 7:400. (2006)
Core transcriptional regulatory circuitry in human hepatocytes
D.T. Odom, R.D. Dowell, E.S. Jacobsen, L. Nekludova, P.A. Rolfe, T.W. Danford, D.K. Gifford, E. Fraenkel, G.I. Bell, and R.A. Young.
Molecular Systems Biology 2:2006.0017. (2006)
High-resolution computational models of genome binding events
Y. Qi, P.A. Rolfe, K.D. MacIsaac, G.K. Gerber, D. Pokholok, J. Zeitlinger, T.W. Danford, R.D. Dowell, E. Fraenkel, T.S. Jaakkola, R.A. Young, and D.K. Gifford.
Nature Biotechnology 24(8):963-70. (2006)
Evaluation of several lightweight stochastic context-free grammars for RNA secondary structure prediction
R.D. Dowell and S.R. Eddy.
BMC Bioinformatics 5:71. (2004)
The distributed annotation system
R.D. Dowell, R.M. Jokerst, A. Day, S.R. Eddy, and L. Stein.
BMC Bioinformatics 2:7(2001)